In the era of big data, efficient data storage and management are critical to the success of both scientific research and industrial applications. HDF5, a hierarchical format for managing experimental data, and TsFile, a modern time-series data storage format, each offer unique strengths and design philosophies. This article takes a deep dive into the origins, use cases, and limitations of HDF5, and explores the similarities and differences between HDF5 and TsFile.
Origins of HDF5
HDF5, short for Hierarchical Data Format version 5, is more than just a file format. It encompasses a full data model, software libraries, and a binary file format designed for storing and managing complex data. HDF5 originated in 1987 and was proposed by the GFTF group at the National Center for Supercomputing Applications (NCSA) in the United States.
The original goal of HDF was to develop an architecture-independent file format capable of meeting the growing need to transfer scientific data across diverse computing platforms at NCSA.
Use Cases
HDF5 has found widespread application in fields such as scientific computing, engineering simulation, and weather forecasting—domains that all require efficient management of experimental data.
Case 1: Scientific Data Storage
In scientific research, there is often a need to store and process complex, multidimensional datasets such as matrices or gridded meteorological data. These datasets typically contain rich metadata and require a storage solution that supports efficient data organization and access.Case 2: Sensor Data from Devices
In industrial monitoring and sensor networks, large volumes of data are generated by various sensors. For example, in the structural health monitoring systems of aerospace agencies, sensor data capturing vibrations, temperature, and other parameters play a crucial role in condition monitoring and fault prediction.
 (Figure 1: The structural health monitoring systems of aerospace agencies)
(Figure 1: The structural health monitoring systems of aerospace agencies)
Case 3: Particle Simulation Data Storage
In particle simulation scenarios, simulation programs can generate vast amounts of experimental data, such as particle trajectories and energy deposition metrics. These data are crucial for understanding physical processes and optimizing simulation parameters. Efficient storage and management systems are essential to handle such workloads.
 (Figure 2: The example of particle simulation scenarios)
(Figure 2: The example of particle simulation scenarios)
Although HDF5 is widely used in scientific computing, a large portion of modern sensor and experimental data is fundamentally time-series in nature. This mismatch between data characteristics and storage models motivates the emergence of specialized time-series formats such as TsFile.
Introduction to TsFile
In many HDF5-based applications, a significant portion of the data actually exhibits time-series characteristics. TsFile is a columnar storage file format specifically designed for time-series data. It was originally developed by the School of Software at Tsinghua University and became a top-level Apache project in 2023.
TsFile stands out with its high performance, high compression ratio, self-describing format, and support for flexible time-range queries.
 (Figure 3: TsFile, a file format for Internet of Things)
(Figure 3: TsFile, a file format for Internet of Things)
Technical Comparison: TsFile vs. HDF5
The following table outlines a technical comparison of TsFile and HDF5 across several key dimensions:
Let’s explore each of these dimensions in more detail:
Compression Ratio
TsFile: TsFile leverages time-series-specific encodings (e.g.,
TS_2DIFFfor timestamp delta encoding,GORILLAfor floating-point compression) along with efficient general-purpose compression algorithms (e.g., Snappy, ZSTD, LZ4). These work in tandem to eliminate redundancy. For variable-length objects, TsFile dynamically allocates memory, avoiding byte-padding waste. Its compact storage strategies make it especially suitable for sparse datasets and variable-length strings.HDF5: HDF5 provides general-purpose compression filters(e.g., gzip, LZF, SZIP) and a plugin mechanism, but lacks built-in time-series–aware encoding schemes such as delta or Gorilla-style compression. As a result, it cannot fully exploit temporal patterns, leading to significantly lower compression ratios. Moreover, for variable-length or sparse sensor data, HDF5 often resorts to fixed-size compound records or multiple small datasets, leading to metadata overhead and padding inefficiencies.
Query & Filtering Capabilities
TsFile: Offers powerful query capabilities, enabling precise reads based on sequence IDs and time ranges. It avoids loading the entire dataset, which significantly boosts query efficiency for large-scale time-series workloads.
HDF5: HDF5 supports partial reads through hyperslabs and chunked storage. However, it lacks native semantic indexing or predicate pushdown for time-based filtering, so efficient time-range queries require external indexing logic.
Data Model
TsFile: Purpose-built for time-series workloads, TsFile adopts a streamlined timestamp–value model. Its schema is simple and well-suited for the sequential and append-heavy nature of time-series data.
HDF5: While highly flexible and capable of modeling complex data structures like multidimensional arrays and compound types, HDF5’s data model is comparatively heavy and not optimized for the unique characteristics of time-series data.
Practical Comparison via Code Examples
While Section 4 presented a conceptual comparison between TsFile and HDF5 across core characteristics such as compression, query flexibility, and data modeling, this section dives into the developer-facing differences through concrete code examples. We demonstrate how both formats handle data writing and querying, and discuss the design implications observed through their APIs. We use the same dataset in both cases: time-series data generated by a device (device1) in a factory (factory1), with the schema (time: long, s1: long).
Example: Writing Data
Example uses TsFile native C++ writer API (standalone TsFile SDK).
TsFile Write Snippet:
// Create a new TsFile named "test.tsfile"
file.create("test.tsfile", O_WRONLY | O_CREAT | O_TRUNC, 0666);
// Define schema for the timeseries table
auto* schema = new storage::TableSchema(
"factory1",
{
common::ColumnSchema("id", common::STRING, common::LZ4, common::PLAIN, common::ColumnCategory::TAG),
common::ColumnSchema("s1", common::INT64, common::LZ4, common::TS_2DIFF, common::ColumnCategory::FIELD),
});
// Create a writer using the schema
auto* writer = new storage::TsFileTableWriter(&file, schema);
// Prepare a tablet for batched insertion
storage::Tablet tablet("factory1", {"id1", "s1"},
{common::STRING, common::INT64},
{common::ColumnCategory::TAG, common::ColumnCategory::FIELD}, 10);
// Insert rows into the tablet
for (int row = 0; row < 5; row++) {
long timestamp = row;
tablet.add_timestamp(row, timestamp);
tablet.add_value(row, "id1", "machine1");
tablet.add_value(row, "s1", static_cast<int64_t>(row));
}
// Write to disk and finalize
writer->write_table(tablet);
writer->flush();
writer->close();HDF5 Write Snippet:
typedef struct {
long time;
long s1;
} Data;
// Create HDF5 file (overwrite if exists)
hid_t file_id = H5Fcreate("test.h5", H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT);
// Create group under root
hid_t group_id = H5Gcreate2(file_id, "factory1", H5P_DEFAULT, H5P_DEFAULT, H5P_DEFAULT);
// Create 1D dataspace
hsize_t dims[1] = { (hsize_t)rows };
hid_t dataspace_id = H5Screate_simple(1, dims, NULL);
// Define compound datatype
hid_t datatype_id = H5Tcreate(H5T_COMPOUND, sizeof(Data));
H5Tinsert(datatype_id, "time", 0, H5T_NATIVE_LONG);
H5Tinsert(datatype_id, "s1", sizeof(long), H5T_NATIVE_LONG);
// Enable chunking and GZIP compression
hid_t dcpl = H5Pcreate(H5P_DATASET_CREATE);
hsize_t chunk_dims[1] = { (hsize_t)rows };
H5Pset_chunk(dcpl, 1, chunk_dims);
H5Pset_deflate(dcpl, 1);
// Create dataset under the group
hid_t dataset_id = H5Dcreate2(group_id, "machine1", datatype_id, dataspace_id, H5P_DEFAULT, dcpl, H5P_DEFAULT);
// Allocate and fill data buffer
Data* dset = (Data*)malloc(rows * sizeof(Data));
for (int i = 0; i < rows; i++) {
dset[i].time = i;
dset[i].s1 = i;
}
// Write and clean up
H5Dwrite(dataset_id, datatype_id, H5S_ALL, H5S_ALL, H5P_DEFAULT, dset);
free(dset);
H5Dclose(dataset_id);
H5Gclose(group_id);
H5Sclose(dataspace_id);
H5Fclose(file_id);Query Example
TsFile Query Snippet:
storage::TsFileReader reader;
reader.open("test.tsfile");
std::vector<std::string> columns = {"id1", "s1"};
storage::ResultSet* result = nullptr;
// Query by table name, columns, and time range
reader.query("factory1", columns, 0, 100, result);
auto* ret = dynamic_cast<storage::TableResultSet*>(result);
bool has_next = false;
while ((code = ret->next(has_next)) == common::E_OK && has_next) {
std::cout << ret->get_value<Timestamp>(1) << std::endl;
std::cout << ret->get_value<int64_t>(1) << std::endl;
}
ret->close();
reader.close();HDF5 Read Snippet:
file_id = H5Fopen("test.h5", H5F_ACC_RDONLY, H5P_DEFAULT);
group_id = H5Gopen2(file_id, "factory1", H5P_DEFAULT);
dataset_id = H5Dopen2(group_id, "machine1", H5P_DEFAULT);
datatype_id = H5Dget_type(dataset_id);
dataspace_id = H5Dget_space(dataset_id);
hsize_t dims[1];
H5Sget_simple_extent_dims(dataspace_id, dims, NULL);
int rows = (int)dims[0];
Data* dset = (Data*)malloc(rows * sizeof(Data));
H5Dread(dataset_id, datatype_id, H5S_ALL, H5S_ALL, H5P_DEFAULT, dset);
for (int i = 0; i < rows; i++) {
printf("Row %d: time: %ld, s1: %ld\n", i, dset[i].time, dset[i].s1);
}
free(dset);
H5Dclose(dataset_id);
H5Gclose(group_id);
H5Sclose(dataspace_id);
H5Fclose(file_id);Interface Behavior in Comparison
Writing: Metadata and Structure
TsFile requires only table name and column types. Its tablet model separates timestamps and values for better compression and performance.
HDF5 demands manual definition of dimensionality and compound types. There is no time-series awareness in structure.
Querying: Filtering and Performance
TsFile supports built-in time-range queries and partial column reads. Filtering is pushed down, reducing unnecessary I/O.
HDF5 offers full dataset reads or chunk-level access. It does not provide built-in predicate pushdown; filtering logic typically runs after data is loaded into application memory.
Developer Experience
TsFile provides intuitive abstractions for time-series data: schema, tablet, time-based queries.
HDF5 exposes low-level controls for flexible data modeling but with significant API complexity.
Real-World Case Study: Aerospace Sensor Data
In a real-world aerospace project, time-series data is primarily collected from aircraft-mounted sensors. Each year, there are thousands of flights, and each flight generates data from approximately 3,000 to 4,000 sensors. These sensors capture a wide range of parameters with varying sampling frequencies and data lengths.
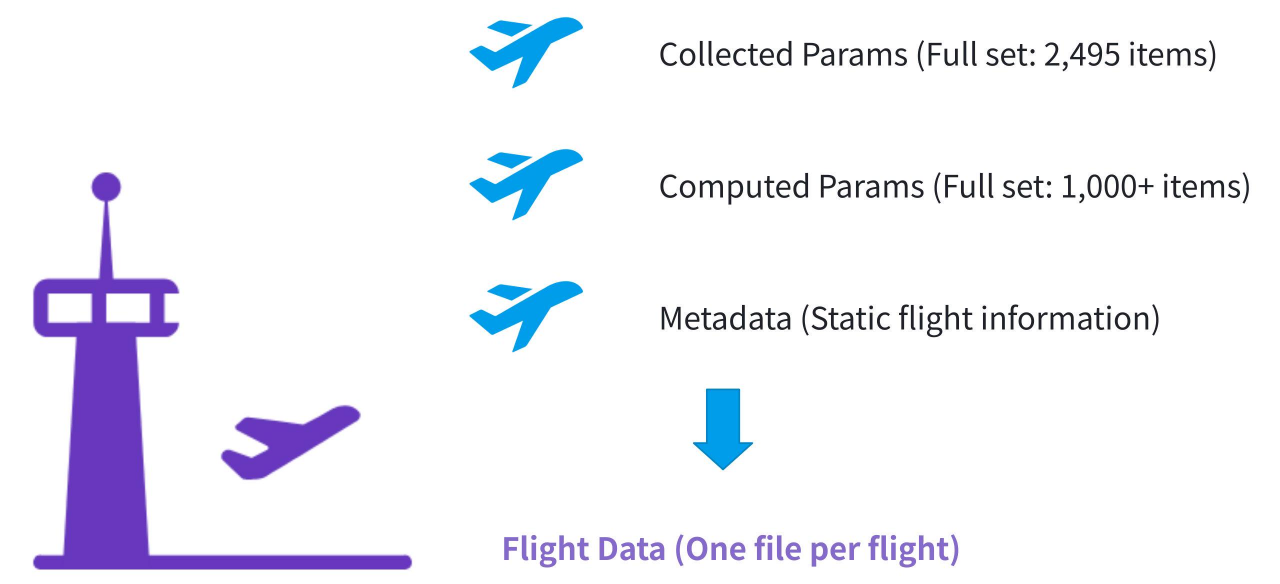 (Figure 4: Aerospace sensor data collection)
(Figure 4: Aerospace sensor data collection)
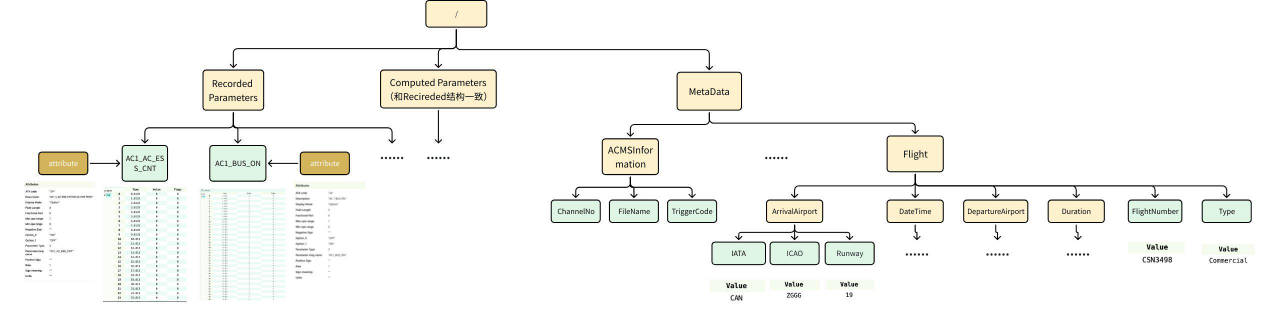
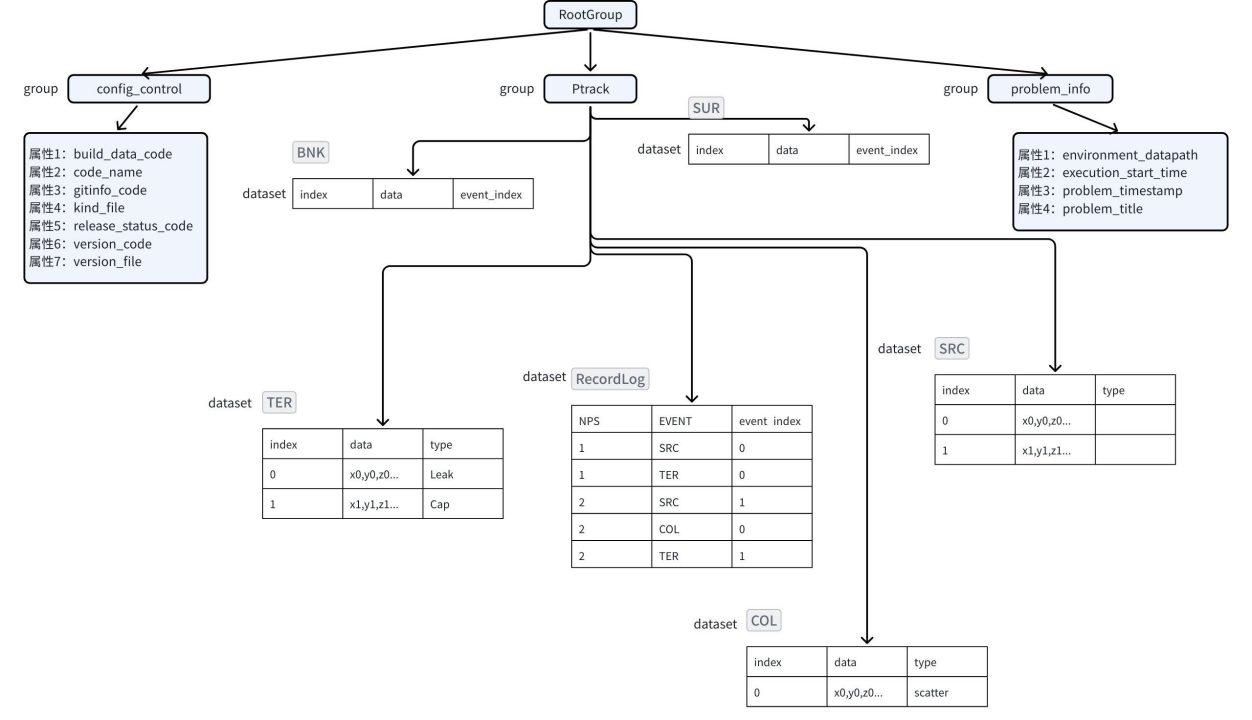
Given the enormous data volume, efficient storage becomes critical. In the HDF5 format, data is organized hierarchically using Groups and Datasets, with metadata stored in Attributes. Each parameter is stored as a separate dataset—a 2D table consisting of a time column and a value column. Internally, HDF5 manages hierarchical objects using internal metadata trees and heaps to map groups to datasets and attributes.
In contrast, TsFile, purpose-built for time-series data, organizes data in a “device–measurement” tree structure. All measurements for a single device are stored contiguously in the file and benefit from columnar compression. Its indexing mechanism consists of a two-level B-tree, linking from the root to devices and then to individual time series. Unlike HDF5, TsFile integrates the time and value columns into a single structure and leverages built-in indexing for fast retrieval without requiring external metadata.
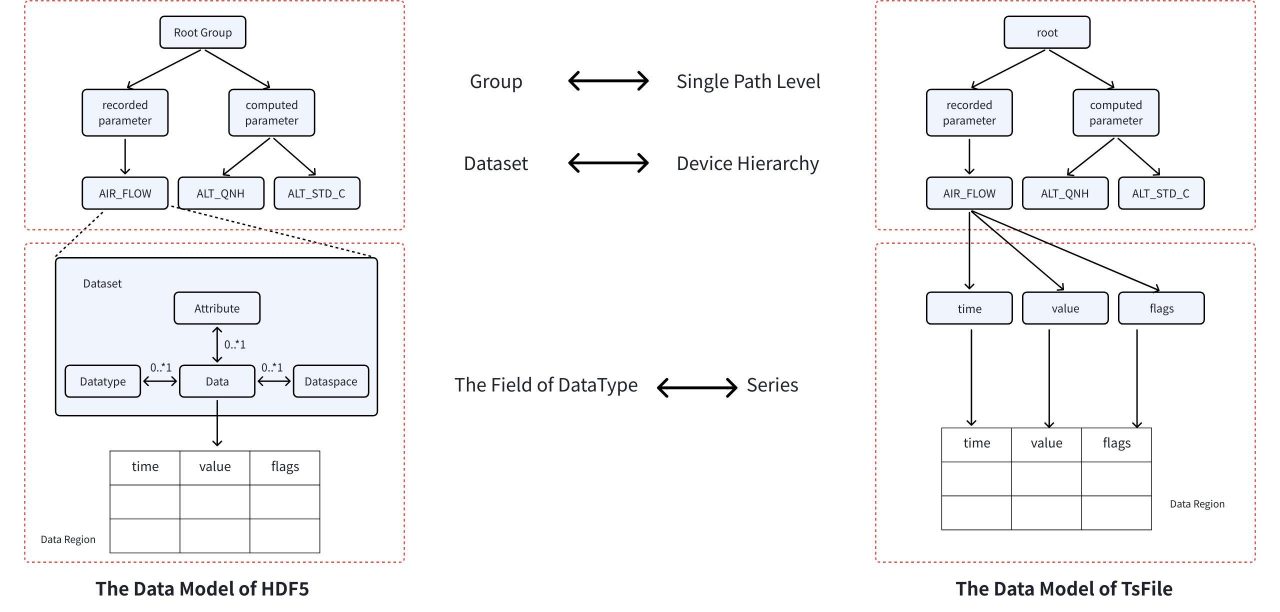 (Figure 5: The Data Model of TsFile and HDF5)
(Figure 5: The Data Model of TsFile and HDF5)
In actual usage, TsFile significantly outperformed HDF5 in both write and query performance. For the same dataset, HDF5 (with compression) occupied approximately 18 TB, using default TsFile encoding (TS_2DIFF + Gorilla + LZ4) and gzip-compressed HDF5 baseline, whereas TsFile (with its native compression and default settings) reduced this to only 2.2 TB—a storage reduction of over 85%. That means TsFile achieved a compression ratio 8 times better than HDF5, with its file size being just 14.31% of the HDF5 equivalent.
Conclusion
With advantages in time-series modeling, compression and encoding schemes, and query filtering capabilities, TsFile proves to be highly optimized for large-scale time-series workloads. The API is also simpler and cleaner, which reduces the learning curve and improves developer efficiency.
Thanks to its performance, storage efficiency, and developer-friendly interface, TsFile stands out as a strong choice for systems that demand high-throughput, low-latency processing of time-series data. For applications requiring scalable and efficient time-series data handling, TsFile is undoubtedly a better fit than general-purpose formats like HDF5.




